SORTIE-ND
Software for spatially-explicit simulation of forest dynamics |
|||||||
Relative Neighborhood Density CalculatorParameters for this behavior
This behavior calculates the relative neighborhood density index (Ω ) as described in Condit et al 2000. Ω is a measure of the aggregation of a species. It is the average density of conspecific neighbors at a certain distance divided by the overall density of trees in the plot. A perfectly random distribution of individuals for a species would result in Ω = 1. Ω > 1 at short distances indicates species aggregation. Ω < 1 at short distances indicates spacing among individuals. How it worksΩ values are calculated for each species for a succession of distances out to a maximum. For each tree, the conspecific neighbors are counted in an ring described by the radii x to Δ x. This value is divided by the area of the ring to get the density of neighbors at that distance. This density is averaged over all trees of each species. This average density of neighbors at a given distance is then relativized by dividing by the density of that species across the plot. 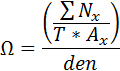 where:
The size of the distance increment, x, is given in the Relative Neighborhood Density - Distance Increment (m) parameter. The maximum distance out to which to calculate Ω is given in the Relative Neighborhood Density - Maximum Distance (m) parameter. Only saplings and adult trees are included in Ω calculations. The resulting Ω values are stored in the Relative Neighborhood Density grid. How to apply itAdd this behavior to your run. There is no need to apply it to specific tree species or types. Indeed, any such specifications will be ignored. This behavior does not automatically create output. Once you have added this behavior to your run, the Detailed output grid setup window will list the Relative Neighborhood Density grid. Save all the data members of this grid. You can then view the Ω values as a line graph and use the graph to save the Ω values as a text file. |
|||||||
|
FAQ - Contact Us
|
|||||||